Introduction
The virus outbreaks of recent years, such as Zika, MERS-CoV, Swine influenza, SARS-CoV, and SARS-CoV-2, illustrate the risk they pose and the intensive research virology requires. Virus assays are used to study viral replication, enzymes, cell entry mechanisms, and many more. In this respect, it is vital to understand various available viral quantification methods to analyze and quantify viruses for research correctly.
Viral quantification methods
Viral quantification is crucial for studying viral biology, replication, and infection mechanisms to accelerate antiviral and vaccine development. Accurate viral concentration determination enables exact normalization of infections and monitoring, optimization, and modification of viral propagation processes to maximize yields. In this article, we briefly describe some of the molecular and immunological approaches for the quantification of viruses along with their advantages and disadvantages (Table 1).

Viral plaque assay (Fig.1) is the most used quantitative viral assay. Plaque assays involve infecting a confluent monolayer of cells with a lytic virus at various dilutions. Infected monolayers are then immobilized with an overlay material. Plaques form as a result of limited infection and replication. Plaques are then counted with neutral red or crystal violet. The virus titer is expressed in plaque-forming units (PFU) per milliliter (PFU/mL).
 Figure 1 Viral Plaque Assay
Figure 1 Viral Plaque Assay
- Median Tissue Culture Infectious Dose (TCID50) is the virus dose required to infect 50% of inoculated cells. The virus is introduced to host cells in successive dilutions. A plate reader and a cell viability assay are then used to assess the proportion of dead cells after incubation.
2. Methods that measure viral protein antigens
- Quantitative Polymerase Chain Reaction (qPCR) (Fig. 2) qPCR measures the quantity of viral DNA or RNA in a sample. PCR is performed with a virus-specific primer in the presence of a dsDNA dye or a sequence-specific reporter. The signal from the reporting fluorophore grows as the DNA amplification proceeds. The number of cycles (Ct value) required to generate a specified fluorescence level is compared to the Ct value of the standard.
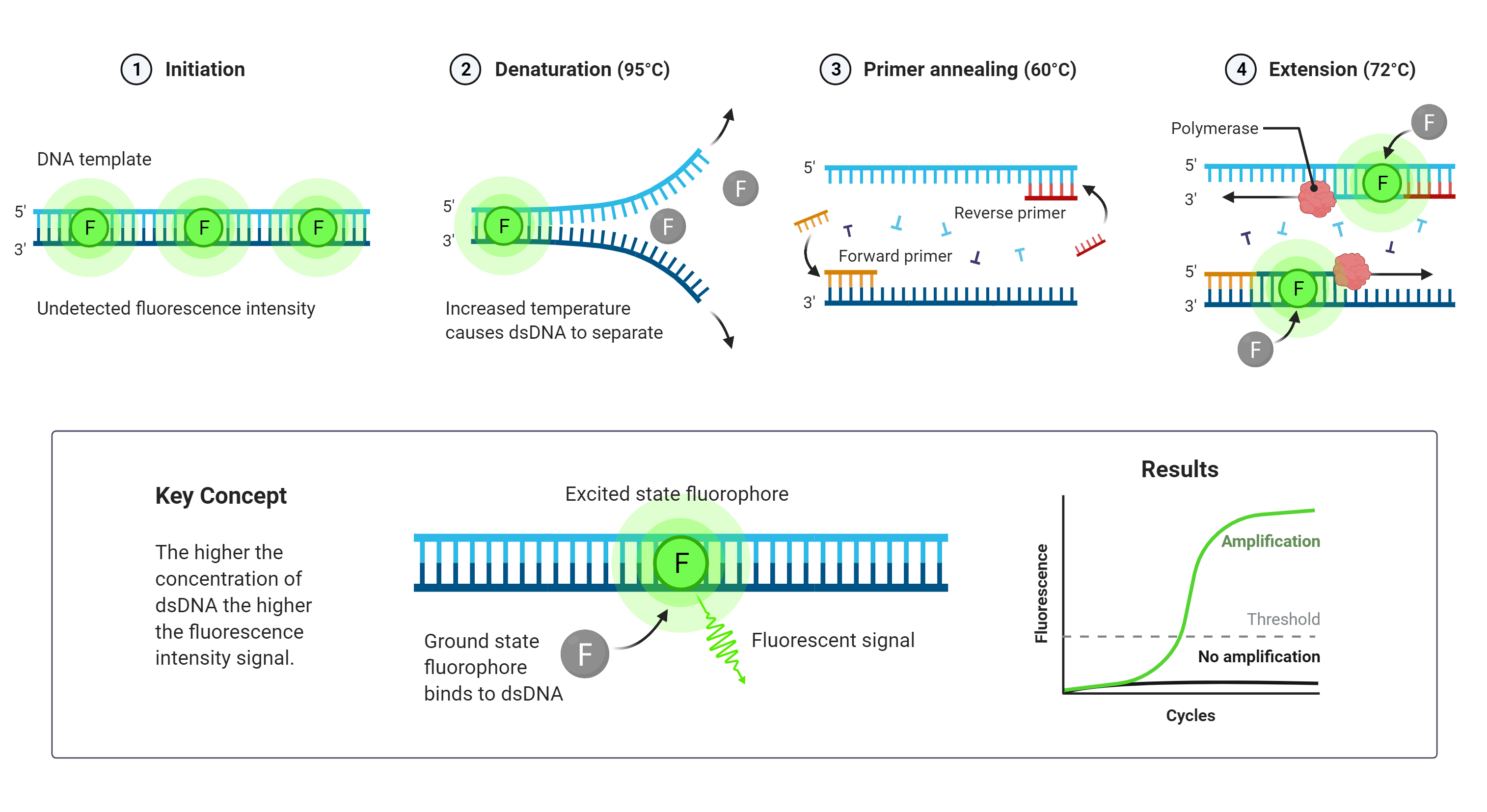
Figure 2 qPCR
- Enzyme-Linked Immunosorbent Assay (ELISA) (Fig. 3) tests serological samples for the presence and/or concentration of antibodies generated by infection or viral antigens. A viral antigen or an antibody against the pathogen is first immobilized in a well which is then recognized by viral antibodies or virus-specific proteins introduced into the well. Antibodies that recognize the attached molecule generate a signal that is measured by a microplate reader. An antigen or antibody signal is proportional to the sample concentration.
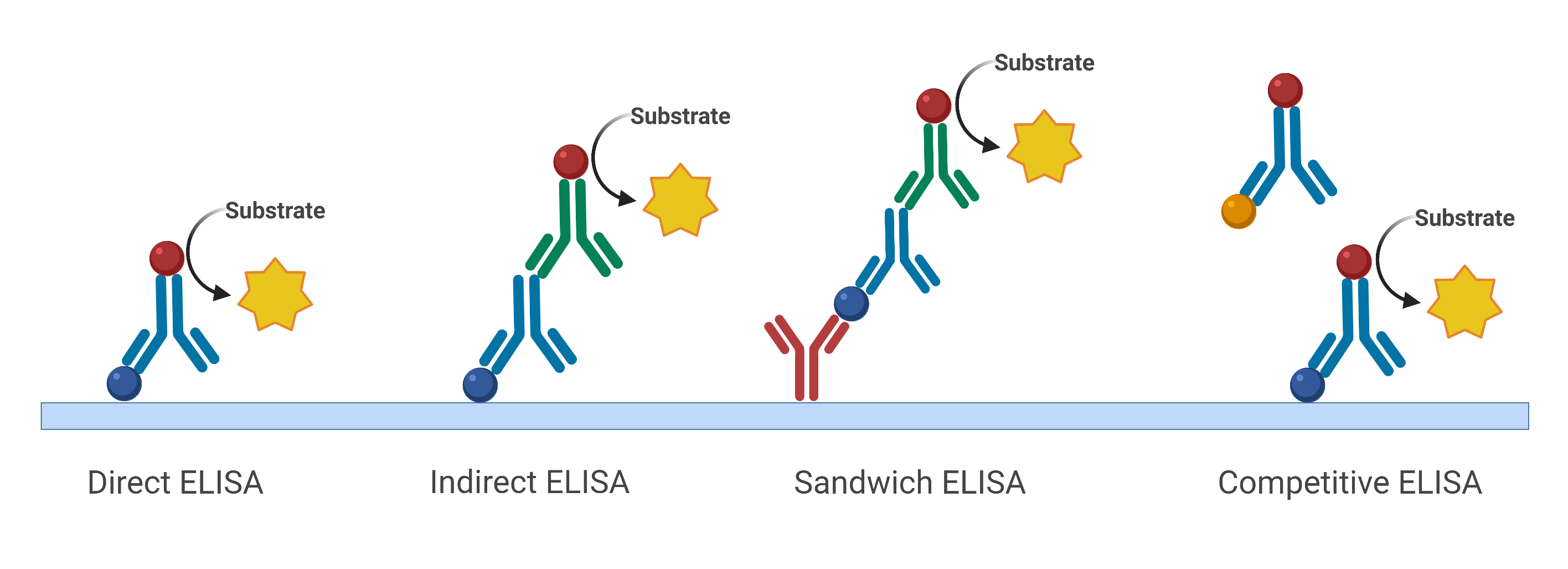 Figure 3 Different types of ELISA
Figure 3 Different types of ELISA
- Hemagglutination Assay (HA) (Fig. 4) assay is used to test influenza viruses' ability to aggregate red blood cells (RBCs). Viral inoculum is diluted and added to mammalian blood samples. Viruses with hemagglutinin protein cluster RBCs into foci of aggregated RBCs, which expand into networks incapable of normal precipitation. Wells are then scored based on whether RBCs precipitate into the wells or not, forming visible spots giving only a relative assessment of viral quantification.

Figure 4 Hemagglutination Assay
3. Methods that directly count viral particles
- Viral Flow Cytometry (Fig. 5) counts intact virus particles in a sample by fluorescently detecting colocalized proteins and nucleic acids. A laser beam analyzes samples labeled with two dyes, one for proteins and one for nucleic acids. In addition to the measured sample flow rate, the number of particles creating simultaneous events on each of the two separate fluorescence channels is determined to calculate the concentration of virus particles per mL.

Figure 5 Viral Flow Cytometry - J Virol. 2018 Jan 17;92(3):e01765-17. doi: 10.1128/JVI.01765-17 (View Source)
4. Methods that optically detect virions by microscopy:
- Immunofluorescence assay is used for the detection of viral antigens or antiviral antibodies in clinical samples. The assay is conducted in two formats: direct immunofluorescence assay (DFA) that detects viral antigens, and indirect immunofluorescence assay (IFA) that detects antiviral antibodies. In the DFA, the antibody that recognizes the viral antigen is directly conjugated to a fluorescent dye. In the IFA, viral antigen-specific antibody is unlabeled and is detected with a second fluorescently labeled anti-human antibody
Nobel Life Sciences (NLS) provides a portfolio of optimized viral quantification assays to screen antivirals, antibodies, and vaccines against many human viruses such as SARS-CoV-2, influenza, and respiratory syncytial virus (RSV). Assays can be chosen and customized to address your specific concerns and needs, ensuring that an appropriate testing plan is designed for your product. NLS is an FDA inspected GLP facility. Our facilities have the capability in BSL3 containment and have been accredited fully and continuously by the AAALAC since 1989.
Please feel free to contact our dedicated team at info@noblelifesci.com
